Biomedical Network Science Lab
Welcome to the Biomedical Network Science Lab!
The Biomedical Network Science (BIONETS) lab investigates molecular disease mechanisms using techniques from network science, combinatorial optimization, and artificial intelligence. We develop algorithms and tools to mine multi-omics data for such mechanisms and to individuate novel strategies for mechanistically grounded drug repurposing and causally effective treatments of complex diseases. We also develop privacy-preserving decentralized biomedical AI solutions, which enable cross-institutional studies on sensitive data. Finally, we are interested in meta-scientific questions such as reproducibility and the impact of data bias on biomedical AI systems.
On July 1, Nicolai Meyerhöfer started as a doctoral researcher in the BIONETS lab. He will work on graph neural network models for prediction of tissue-specific protein-protein interactions.
In our joint retreat with the DaiSyBio group (TUM) in Peschiera del Garda, we had inspiring talks and discussions on hot topics and off topics in computational systems biology, ran two hackathons, and engaged in a series of team building events.
In our paper "Efficiently Labeling and Retrieving Temporal Anomalies in Relational Databases", we show how to flag and retrieve temporal inconsistencies in healthcare information systems.
In our paper "Drugst.One — a plug-and-play solution for online systems medicine and network-based drug repurposing", we present a versatile plugin to turn any tool that outputs a list of genes or proteins into a feature rich, drug repurposing web tool with interactive network visualization.
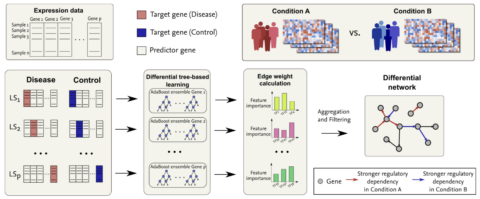
In our paper "Inference of differential gene regulatory networks using boosted differential trees", we present BoostDiff, a tree-based method for identifying differences in transcriptional regulation between two conditions.